News & Press
How Microparticles Come Together
Researchers develop new method to translate molecular dynamic structures to the microscopic level
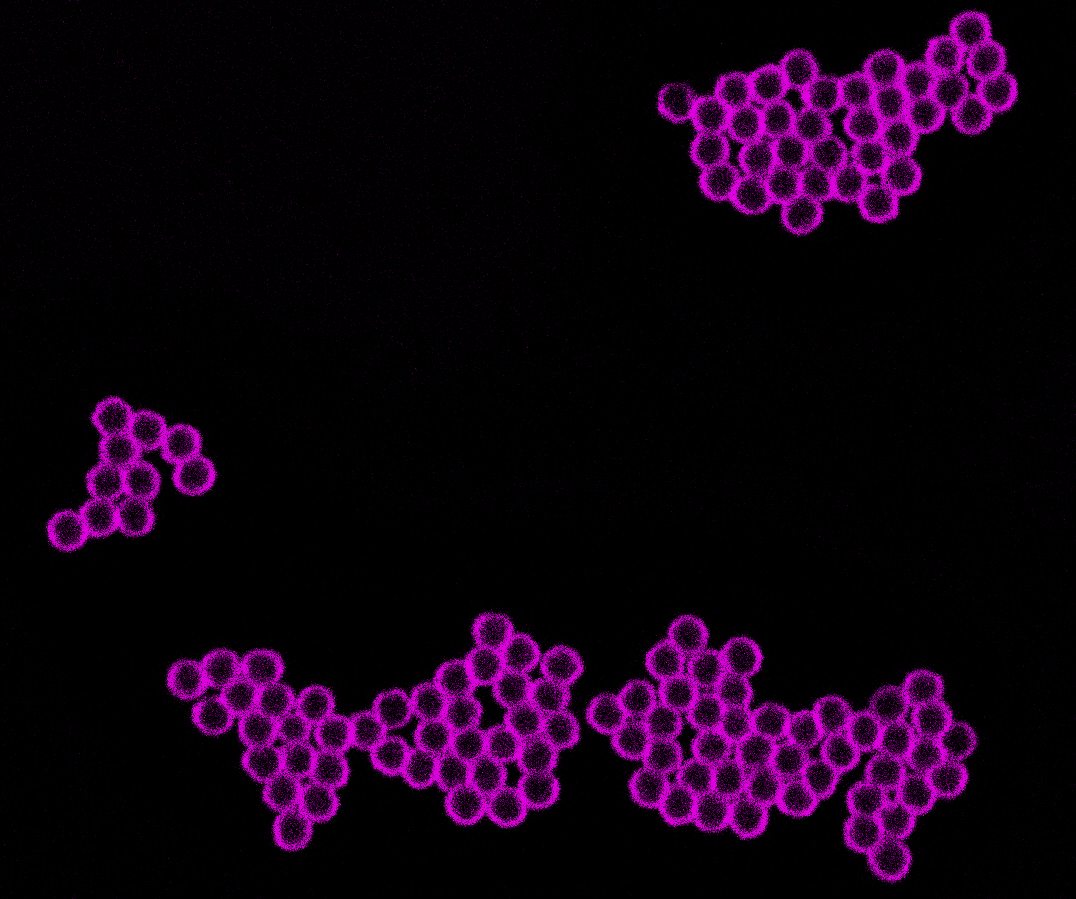
Multiple Colloids form a larger self-organized structure after the polymerization of the DNA. Source: Jie Deng
Molecular structures such as microtubules control essential functions in the cell, such as cell division or locomotion. The scaffolding structure, which occurs in cells of plants, animals and humans, is composed of proteins and is constantly being built and degraded dynamically and simultaneously. This enables the structure to react quickly to stimuli and adapt to external conditions. One goal in materials science is to develop synthetic molecular structures that are as adaptable as microtubules. They could provide the basis for material systems that regulate themselves. However, it has not yet been possible to transfer such dynamic systems from the molecular level to the material level.
Prof. Andreas Walther from the Cluster of Excellence “Living, Adaptive and Energy-autonomous Materials Systems” (livMatS) and Jie Deng from the Institute of Macromolecular Chemistry at the University of Freiburg have presented a new method that represents a first step in this direction. With this method, researchers can transfer the molecular self-assembly of DNA building blocks across hierarchical length scales and thus control the self-assembly of microparticles. The researchers have published their results in the journal Nature Communications.
In order for different building blocks to come together in the cell to form larger structures such as microtubules, biomolecular recognition is required. For example, proteins have characteristic sequences to which a compatible substance can dock. The new method uses the principle of molecular recognition: The Freiburg researchers use the enzyme T4 DNA ligase to connect individual DNA monomers to form long DNA polymers. For this, the enzyme needs adenosine triphosphate as an energy supplier. The enzyme BsaI, which can recognize and cut DNA at specific positions, cleaves the binding sites. This causes the polymers to form and degrade simultaneously. BsaI cuts the DNA-strands downstream its recognition site and does not prefer any particular sequence. Thus, the system allows flexible connection of different DNA building blocks in a predesigned order.
This feature can be used to bring functional side chains, for example molecules, into contact with each other: By attaching them to the ends of the DNA-strands, they can interact with each other. Deng and Walther were able to further program the side chains to bind to single-stranded DNA attached to the surface of a microparticle, a so-called colloid. After the polymerization, the DNA can wrap among multiple colloids. This leads to the colloids coming together in larger self-organized structures. With their method, the researchers can form structures consisting of two different types of DNA polymers and self-assembling colloids, in which the two types of colloids aggregate separately.
“As such, we are able to achieve dynamic systems a step closer to natural structures”, says Jie Deng. “The systems may be further used to design dynamic materials that can interact with cells to regulate biofunctions.”
Original Publication:
Jie Deng, Andreas Walther: ATP-powered molecular recognition to engineer transient multivalency and self-sorting 4D hierarchical systems. In: Nature Communications 11, 3658 (2020), doi: 10.1038/s41467-020-17479-9

Nature Nanotechnology Volume 15 Issue 11, November 2020.
Image: Johannes Richers.
Cover Design: Bethany Vukomanovic.
Edit 05 November 2020
An artist's interpretation of this research is depicted on the cover of the November 2020 issue of the journal Nature Nanotechnology.
